Improving Accuracy in Chemical Simulation
Chemical simulation is an essential tool in the physical sciences. Offering an atomistic, or even electronic, insight into the relevant systems of interest. However, chemical simulation is computationally expensive. With growing economic and environmental costs associated with compute, it is necessary that the chemical simulation that is run is analysed so that the maximum information is obtained from the minimum simulation.
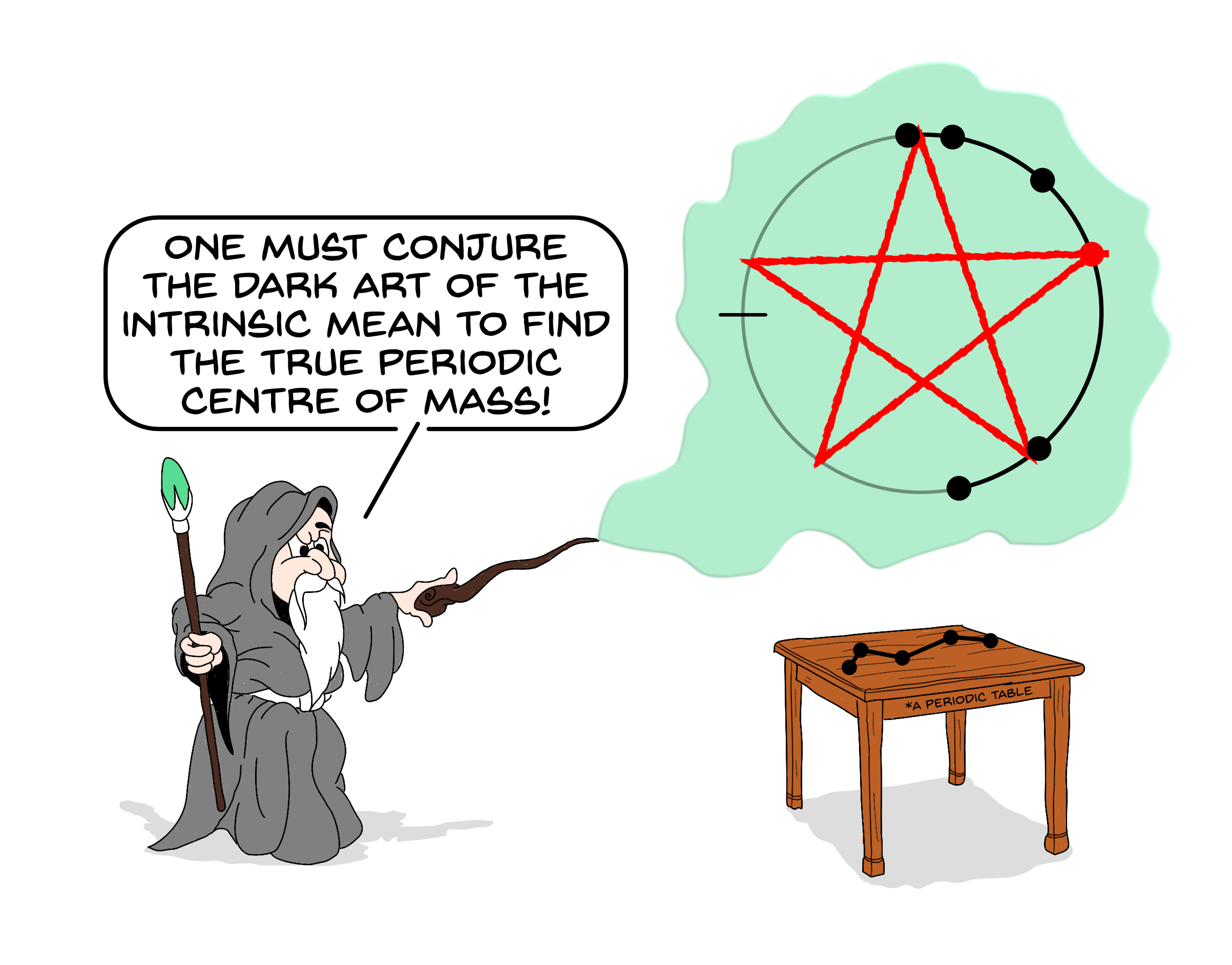
To address this problem, we look to improve the analysis methods applied to simulation data. This work has included the development of a statistically efficient Bayesian regression framework for diffusion coefficient estimation,1 which we have implemented in the open-source Python package kinisi.2 Recently, we have addressed the problem of estimating the centre of mass of a molecule in a periodically repeating cell. Specifically, we have highlighted the need to compute the intrinsic centre of mass in the circular periodic cell, rather than the extrinsic centre of mass that is popularly used3.
Relevant Publications
-
A. R. McCluskey, S. W. Coles, & B. J. Morgan. J. Chem. Theory Comput., 21(1), 79, 2025. DOI: 10.1088/2632-2153/ab94c4. ↩
-
A. R. McCluskey, A. G. Squires, J. Dunn, S. W. Coles, B. J. Morgan. J. Open Source Softw., 9(94), 5984, 2024. DOI: 10.21105/joss.05984 ↩
-
H. Richardson, J. Dunn, & A. R. McCluskey. arXiv:2501.14578, 2025. ↩